08. Basic DICOM Volume EDA
Basic DICOM volume EDA Heading
Exploratory Data Analysis of DICOM datasets
Now let me walk you through a Python Notebook where I analyze a single 3D medical image volume. This notebook and the volume used by it are available among the course materials.
ND320 C3 L2 07 Basic DICOM Volume EDA
Video Summary
We have just seen a few basic techniques for working with medical imaging volumes. We have hit on a few quite important things to remember when exploring your data and thinking of preparing volumes for downstream machine learning pipelines. You are welcome to explore the Notebook on your own at the bottom of the page.
Voxel spacing
DICOM voxels do not have to be perfect cubes (as they are in many computer vision problems). There are DICOM Data Elements that will tell you what exactly are the dimensions of voxels. The most important ones are Pixel Spacing and Slice Thickness. However, there are others, and if your project involves measuring things, make sure you get the transformation right by closely inspecting the tags in your dataset and comparing them with the list of elements in the IOD table for the respective modality.
Data ranges
We have seen how with CT, you may have data in your dataset that will represent synthetic material or items artificially added by scanners. It is always a good idea to see if there is something outstanding in the image you are dealing with and if it represents something that you need to think about in your downstream processing.
Conversions between DICOM values and screen space are particularly important if you are planning to visualize slices for any kind of diagnostic use or overlay them on top of diagnostic information. We have not really touched the aspects of visualization other than being mindful of bit depth and doing our own windowing, but DICOM images contain quite a lot of information that defines how exactly you are expected to map the data to the screen colorspace. If you are interested in exploring this further or need to accurately represent the data, take a closer look at elements in DICOM’s ImagePixel module. Things like Pixel Representation, Photometric Interpretation, Rescale Slope, Rescale Intercept and many others define how values should be transformed for accurate representation.
Check for Understanding
SOLUTION:
- (0028,0101) Bits Stored
- (0028,0004) Photometric Interpretation
Quiz - Axial CT Slice
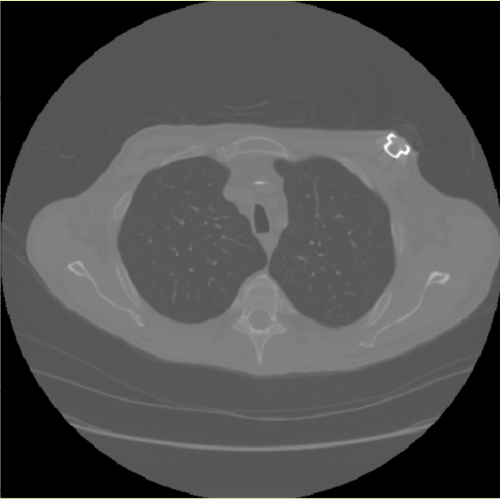
Axial CT slice
Why is CT image round?
QUESTION: Reflect
In the image above, we can see an example of a rendering of an axial CT slice that we encountered in our EDA exercise.
We have talked about why pixels in the corners of this image are black.
Use the textbox below to answer the following question: why are we seeing a distinct circular shape around the patient anatomy in this image?
ANSWER:
Thanks for your response. I hope you guessed quickly that the answer has to do with what we have discussed in Lesson 1. CT scanners acquire data by rotating the x-ray source inside the gantry - hence the circular shape of the acquired image.
WS intro
Below you can find the Jupyter Notebook that was used in this walkthrough.
Code
If you need a code on the https://github.com/udacity.